Remember our friend Mary? She was diagnosed with Canavan Disease when she was an infant, and suffers seizures as a result. To help treat these seizures, she’s been prescribed several different anti-epileptic drugs. Unfortunately, her body has reacted poorly with these medications, and she’s developed hypersensitivity to several different drugs. Originally, she developed rashes and eosinophilia, a higher than normal level of a type of white blood cell. She’s developed fevers, flu-like symptoms, and terrible blisters; which her doctor has diagnosed as Stevens-Johnson syndrome. Due to the severity of this syndrome, medications must be withdrawn and replaced with medications that she hopefully will not react to. She also is anemic and has blood in her urine, which is not attributable to either Canavan Disease or Stevens-Johnson syndrome.
Stevens-Johnson syndrome is associated with mutations in HLA-B, and some diseases are associated with increased risk of this syndrome. Her clinical team has been unable to identify another disease(s) or disorder(s) that may be causing her symptoms. To address her hypersensitivities, anemia, and the blood in her urine, her doctor recommends genetic sequencing. The options they discuss could include sequencing of either the exome or the genome.
Stevens-Johnson syndrome in OMIM
OMIM #608579
Genome: The genome is all of our DNA, including the regions that are never translated into proteins. Mutations in non-coding regions can still be responsible for disease.
Exome: The exome is the part of the genome that is actually expressed as proteins. It only covers ~1.5% of our genome, but mutations in the exome are more likely to cause disease. Whole exome sequencing can identify a diagnosis 25-30% of the time.(Vissers et al, 2016,Seidelmann et al, 2017)
There are several options that they might consider.
- A clinical study is offered by an academic group who is interested in the genetic causes of Stevens-Johnson syndrome.
- Pros: As part of this study, a full-exome sequence would be conducted free of charge and Mary and her family would have free access to the results. Additionally, the analysis would be conducted by geneticists and any potential diagnoses would be revealed.
- Cons: As with any clinical trial, there are also risks, including the potential exposure of her genetic and medical history. In some clinical trials, there are also medical risks, such as when a new drug is introduced.
- Without enrolling in the clinical trial, they could seek coverage of genome analysis from their insurance provider
- Pros: Given the severity of her conditions and with support from her physicians, Mary’s parents could make the case that genome analysis would save the insurance company long-term, potentially avoiding multiple hospital visits and tests. Additionally, since whole-genome sequencing is approximately the same cost of exome sequencing, they would potentially have a greater chance of finding a diagnosis.
- Cons: Negotiations can be timely, taking up to 6 months.
- They could pay for a full genome analysis out of pocket.
- Pros: As any of us who has dealt with negotiating insurance coverage can understand, this might certainly be a less stressful option, if cost is not a concern. Given the already shortened life-expectancy of a child with Canavan disease, expedience is also preeminent in Mary’s parents thoughts. They would get whole-genome analysis as opposed to only exome analysis.
- Cons: It costs 5-10 thousand dollars, out-of-pocket.
- The final option is to use direct-to-consumer options, and then perform an analysis of the raw results.
- Pros: These tests can be done at home. They require no preparation or appointments. They also tend to be fairly cheap.
- Cons: Unless Mary’s parents have extensive scientific training (or are extremely driven), this is an unlikely option. Understanding the data and analyzing it with enough confidence to make a clinical decision is not only difficult, but time consuming, if methods are not already in place. This is especially the case since an entire genome would need to be parsed through. Another potential problem with direct-to-consumer option is the potential for errors in sample attainment and handling.
With any of these options, it is most beneficial if the parents are also sequenced. Mutations in the healthy parent that are also seen in the child are likely not useful hits for diagnosis. However, if both parents are carriers of a mutation (heterozygous), but the child is homozygous for that mutation, the disease may present itself. “Trio” anlalysis (mom, dad, and child) increases diagnostic yield from 20% to 40% in the Centogene platform.
Additionally, complex diseases or phenotypes (e.g. type 2 diabetes, multiple sclerosis, etc.) are not associated with variation in a single locus and WGS cannot provide diagnostic criteria. However, it can provide someone with a risk for a particular diagnosis (and a high risk profile with the corresponding clinical features may provide confidence in a diagnosis).
Since Mary is a child and her condition necessitates aggressive care, it would be beneficial to condict this analysis as quickly as possible. I would therefore not recommend negotiating with their insurance company for coverage. Assuming that her parents do not have training in genetics, I would not recommend that they attempt to analyze the raw data themselves. The least risky option would be to pay for the genome analysis out of pocket. However, Canavan Disease is a chronic condition that requires constant care. This is likely cost-prohibitive for the average family.
I would recommend participating in the clinical trial. As a scientist, this is an easy choice for me to make. Generally clinical trials provide the patient with more information and more close supervision than they would normally receive and it benefits the greater good. However, as a family member of someone with a chronic and debilitating illness, I understand a patient’s reluctance to engage in additional doctor’s visits and tests. Additionally, while clinical trials are designed to be as safe as possible, there are always risks to an experimental treatment. The risks vary by clinical trial, and are sometimes as small as the (very minor) possibility of infection from a needle stick. However, experimental treatments draw larger risks, such as unknown toxicity.
Incidental findings: With any of these options, it is possible that a genetic variant that incurs some risk of a disease that is not related to the current clinical presentation is identified. Ideally, this possibility and the desires should be discussed before any analysis occurs. Remember that House episode where “13” knows that there is a possibility of her inheriting Huntington’s Disease? Although House wanted her to take a genetic test to find out if she had the mutation, she did not want to know because little could be done about the condition. Every person will feel differently about their certain and uncertain risks and a discussion is extremely important.
However, assuming that there is a clear risk or indication of disease, the physician should discuss this with the patients and send them to a specialist who can more closely monitor and diagnose the case. The American College of Medical Genetics has identified more than 50 genes whose variants should be reported even if these genes were not the subject of the initial analysis.Complex diseases are… well… more complex. Incidental findings that indicate a high risk of incidental disease should be discussed to determine any lifestyle choices that can help prevent an outcome. However, the physician should not confer unnecessary worry on a patient or their family. When a minor risk is identified, some patients may take unnecessary steps, such as “preventative” mastectomies. This was one reason that the FDA began regulating 23andme.
Mary’s parents received a VCF formatted file with her sequencing results. Unfortunately it appears the results only confirmed what they already knew: a mutation in ASPA. This is what they’ve received:
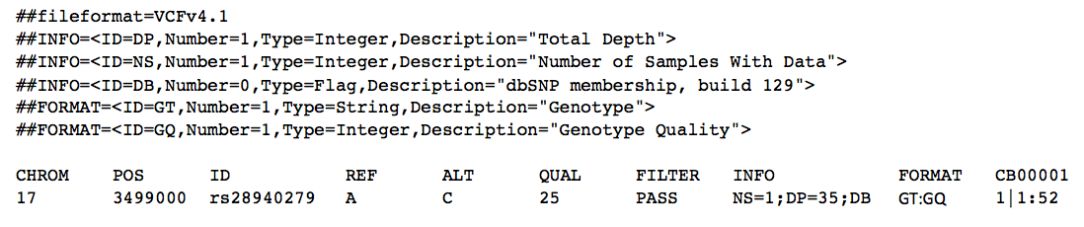
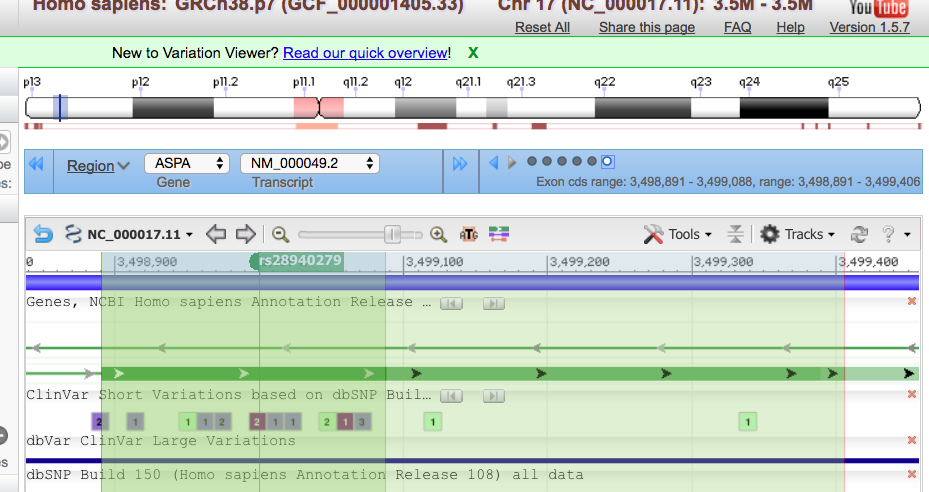
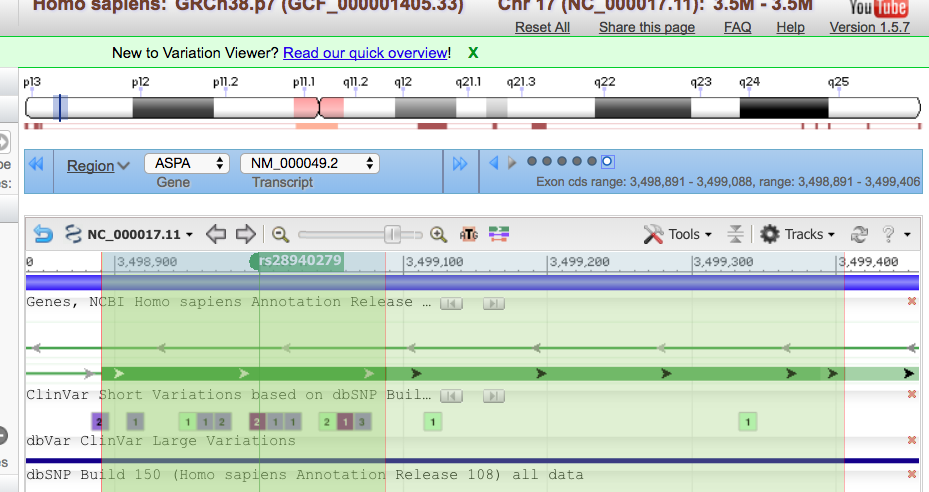
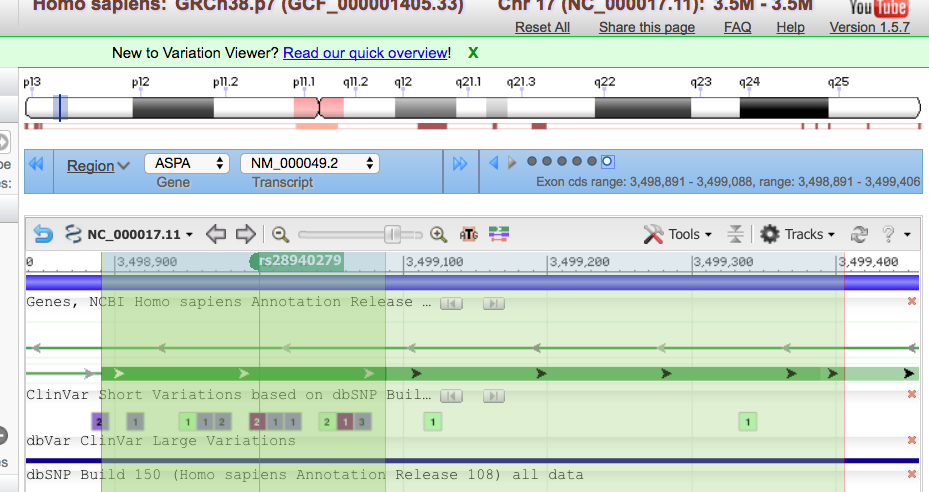
This tells them that she has a variant in chromosome 17, position 3,499,000 (of that chromosome). She has a SNP that has the rsID of 28940279. The reference nucleotide is an A (adenine), and the variant expressed is a C (cytosine) . The quality for the variant is 25, and has passed any quality filters put in place. There is one sample; a combined read depth of 35, and this variant is only found in dbSNP. We have evaluated genotype and genotype quality. The last column is her sample, CB00001. It shows that she is homozygous for the alternate/variant allele and that the quality of that genotype is 52.
From OMIM, we can see that the rdID matches Mary’s mutation, E285A
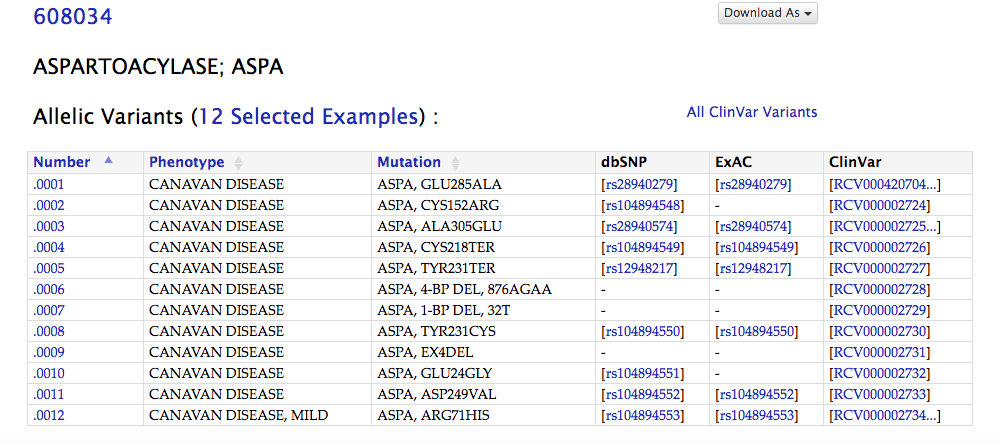
In Varview, we can see that this SNP is in the exon whose coding sequence spans from 3,498-891-3,499,088. This is shown in the blue box.